Discovery of PCR
PCR stands for Polymerase Chain Reaction. It's a widely used molecular biology technique that allows scientists to amplify (make many copies of) a specific DNA segment.
PCR was invented in 1983 by Kary Mullis at Cetus Corporation. Mullis was working to improve the synthesis of oligonucleotides when he envisioned the concept of PCR, which revolutionized the field of molecular biology by enabling rapid DNA amplification. Mullis shared the 1993 Nobel Prize in Chemistry with Michael Smith for his invention.
Components of PCR
The PCR is a method of in-vitro DNA synthesis. This includes the template to be copied, primers to primers synthesis of the template, nucleotides, polymerase enzyme and buffer components monovalent and divalent cations to provide optimal conditions for accurate and efficient replication.
- Primers :- primers determine the specificity of the PCR. Primers are chemically manufactured on a DNA synthesizer. Primers are designed to contain sequences homologous to sites flanking the region to be analysed. Primers are single- stranded DNA fragments, usually 20-30 bases in length. The forward primer must bind to the target DNA sequence just 5' to the sequence intended to be amplified. The reverse primer must bind just 5' to the sequence to be amplified on the opposite strand of the DNA.
- Primers dimers :- these are PCR products that are just double the size of the primers.
- DNA Template:- the DNA sample containing the target sequence to be amplified. The template may be single or double stranded DNA from the patient's genomic or mitochondrial DNA, viruses, bacteria, fungi or parasites that might be infecting the patient.
- dNTPs :- deoxynucleotide Triphosphates, the building blocks for new DNA strands dATP, dTTP, dCTP, dGTP.
- DNA polymerase :- an enzyme that synthesizes new DNA strands by extending the primers. Commonly used polymerases include Taq polymerase.
- Buffer solution :- maintains the optimal pH & ionic strength for the DNA polymerase to function effectively. It often contains magnesium ions (Mg2+), which are essential for enzyme activity.
- Nuclease-Free-Water :- used to make up the final volume of the reaction mix and ensure no contamination.
Control of PCR contamination
- Contamination is a significant concern for methods that involve target amplification by PCR. Although genomic DNA is a source of spurious PCR target, the major cause of contamination is PCR products from previous amplifications. Contamination is controlled both physically and chemically.
- Ultraviolet (UV) light has been used to decontaminate and maintain pre-PCR area.
- Isolation cabinet are equipped with UV light source that are turned on for about 20 minutes after the box has been used.
- Bleach is a widely used method for decontamination and preparation. Frequently wiping bench tops, tools or any surface that comes in contact with specimen material with dilute bleach or alcohol removes most DNA contamination.
- Another widely used chemical method of contamination control is the dUTP-UNG system. A short incubation period is added to the beginning of the PCR amplification program, usually at 50°c for 2-10 minutes to allow the UNG enzyme to function.
PCR product clean up
PCR product clean up is an essential step to remove unwanted components such as primers, unincorporated dNTPs, enzymes, and salts from the amplified DNA.
Below are common methods for PCR product cleanup:-
1. Enzymatic Cleanup.
Steps:-
a. Add ExoSAP (a mix of Exonuclease 1 and SAP) to the PCR reaction.
b. Incubate at 37°c for 15-30 minutes to allow digestion.
c. Heat inactive the enzymes at 80°c for 15 minutes.
Pros:- Simple and requires minimal handling.
Cons:- Not suitable for larger-scale clean up.
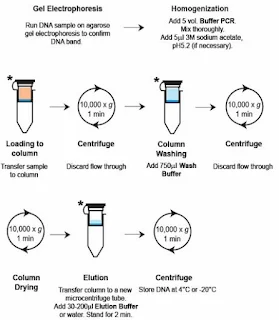
2.Spin Column-Based Purification.
Steps:-
a. Add binding buffer to the PCR product and transfer it to the column.
b. Centrifuge to bind DNA to the silica membrane.
c. Wash with wash buffer to remove impurities.
d. Elute the purified DNA in water or buffer.
Pros:- High purity and yield; scalable for multiple reactions.
Cons:- Requires specialized kits.
3. Magnetic Bead-Based Purification.
Steps:-
a. Mix PCR product with bead-binding buffer and magnetic beads.
b. Incubate and use a magnetic separator to retain beads while removing supernatant.
c. Wash beads and elute purified DNA.
4. Gel Extraction.
Steps:-
a. Run the PCR product on an agarose gel.
b. Excise the desired band under UV light.
c. Extract DNA using a gel extraction kit.
Pros:- Ensures recovery of the correct- sized product.
Cons:- Time-consuming and may result in lower yields.
5. Ethanol Precipitation.
Steps:-
a. Add 2.5 volumes of cold ethanol and 0.1 volume of sodium acetate to the PCR product.
b. Incubate at -20°c for 30 minutes.
c. Centrifuge to pellet the DNA, wash with 70% ethanol, and resuspend in water or buffer.
Pros:- cost-effective.
Cons:- less efficient for small DNA fragments.
What is PCR mispriming? How can it be controlled?
PCR mispriming occurs when primers bind to non- target sequences of the DNA template due to partial or weak complementarity. This results in non-specific amplification, which can lead to undesirable PCR products and reduce the efficiency and specificity of the reaction.
★ Controlling PCR Mispriming:-
- Optimize Primer Design:- Ensure primes are 18-25 nucleotides long with a melting temperature of 55-65°c. Avoid complementarity between primers.
- Adjust Annealing temperature:- perform a gradient PCR to determine the optimal annealing temperature.
- Hot-Start PCR:- use a heat-activated DNA polymerase that remains inactives at room temperature. Prevents non-specific primer binding before thermal cycling begins.
- Optimize Magnesium ion (Mg2+) concentration that supports efficient amplification.
- Touchdown PCR:- start with a high annealing temperature and gradually decrease it during initial cycles
- Use Additives:- like DMSO, formamide, reduce secondary structures and improve primer binding specificity.
- Perform Nested PCR:- use a second round of amplification with internal primers that target a specific region within the initial PCR product.